Note
Click here to download the full example code
EEG without events or epochs
import mne
from mkpy.io import mkh5mne
print(mne.__version__)
mne.viz.set_browser_backend("matplotlib") # for docs generation
Out:
1.0.3
Using matplotlib as 2D backend.
This example is a resting EEG recording with no mkpy codemapped events or epochs tables.
eeg_h5_f = "../mkh5_data/sub000eeg.h5"
mne_raw = mkh5mne.from_mkh5(
eeg_h5_f,
dblock_paths=["open/dblock_0", "closed/dblock_0"],
)
Out:
ok
checking info, montage closed/dblock_0
Creating RawArray with float64 data, n_channels=39, n_times=45568
Range : 0 ... 45567 = 0.000 ... 182.268 secs
Ready.
/usr/share/miniconda/envs/env_3.8/lib/python3.8/site-packages/mkpy/io/mkh5mne.py:1478: RuntimeWarning: Not setting positions of 5 eog channels found in montage:
['lle', 'lhz', 'HEOG', 'rle', 'rhz']
Consider setting the channel types to be of EEG/sEEG/ECoG/DBS/fNIRS using inst.set_channel_types before calling inst.set_montage, or omit these channels when creating your montage.
raw_dblock.set_montage(montage)
Creating RawArray with float64 data, n_channels=39, n_times=45568
Range : 0 ... 45567 = 0.000 ... 182.268 secs
Ready.
/usr/share/miniconda/envs/env_3.8/lib/python3.8/site-packages/mkpy/io/mkh5mne.py:1478: RuntimeWarning: Not setting positions of 5 eog channels found in montage:
['lle', 'lhz', 'HEOG', 'rle', 'rhz']
Consider setting the channel types to be of EEG/sEEG/ECoG/DBS/fNIRS using inst.set_channel_types before calling inst.set_montage, or omit these channels when creating your montage.
raw_dblock.set_montage(montage)
EEG data marked as already having the desired reference.
mne_raw.plot(
scalings={"eeg": 5e-5, "eog": 1e-4},
start=282.0,
n_channels=39,
)
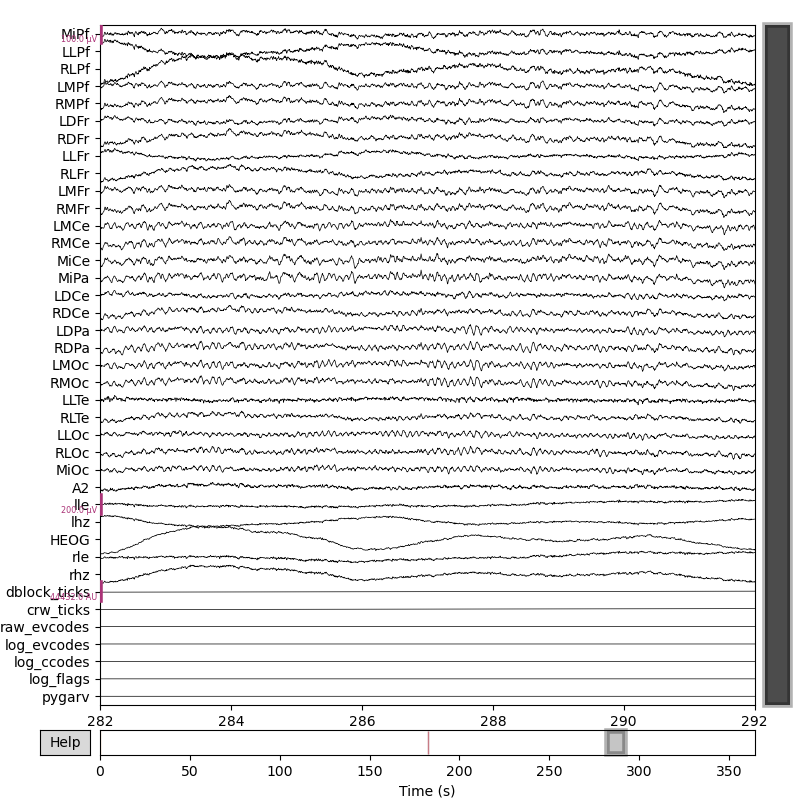
Out:
Opening raw-browser...
<MNEBrowseFigure size 800x800 with 4 Axes>
Total running time of the script: ( 0 minutes 2.563 seconds)