Note
Click here to download the full example code
mkh5 Quickstart
In a terminal window use
mamba(recommended) orconda(if necessary) to install mkpy along with jupyter or jupyterlab into a fresh environment and activate it:$ mamba create --name mkpy_041221 mkpy jupyterlab \ -c kutaslab -c conda-forge -c defaults \ --strict-channel-priority $ conda activate mkpy_041221
In a terminal window navigate to the directory where you want to work and run:
$ jupyter notebook
Open a notebook with a Python 3 kernel and proceed.
Single subject single session workflow
from mkpy import mkh5 as mkh5
DATA_H5_F = "../mkh5_data/_sub000p3.h5"
data_h5 = mkh5.mkh5(DATA_H5_F)
data_h5.reset_all()
# load the crw, log, and YAML header
data_h5.create_mkdata(
"sub000",
"../mkdig_data/sub000p3.crw",
"../mkdig_data/sub000p3.x.log",
"../mkdig_data/sub000p3.yhdr",
)
If calibration pulses are not in the .crw file, append them like so:
data_h5.append_mkdata(
"sub000",
"../mkdig_data/sub000c.crw",
"../mkdig_data/sub000c.log",
"../mkdig_data/sub000c.yhdr",
)
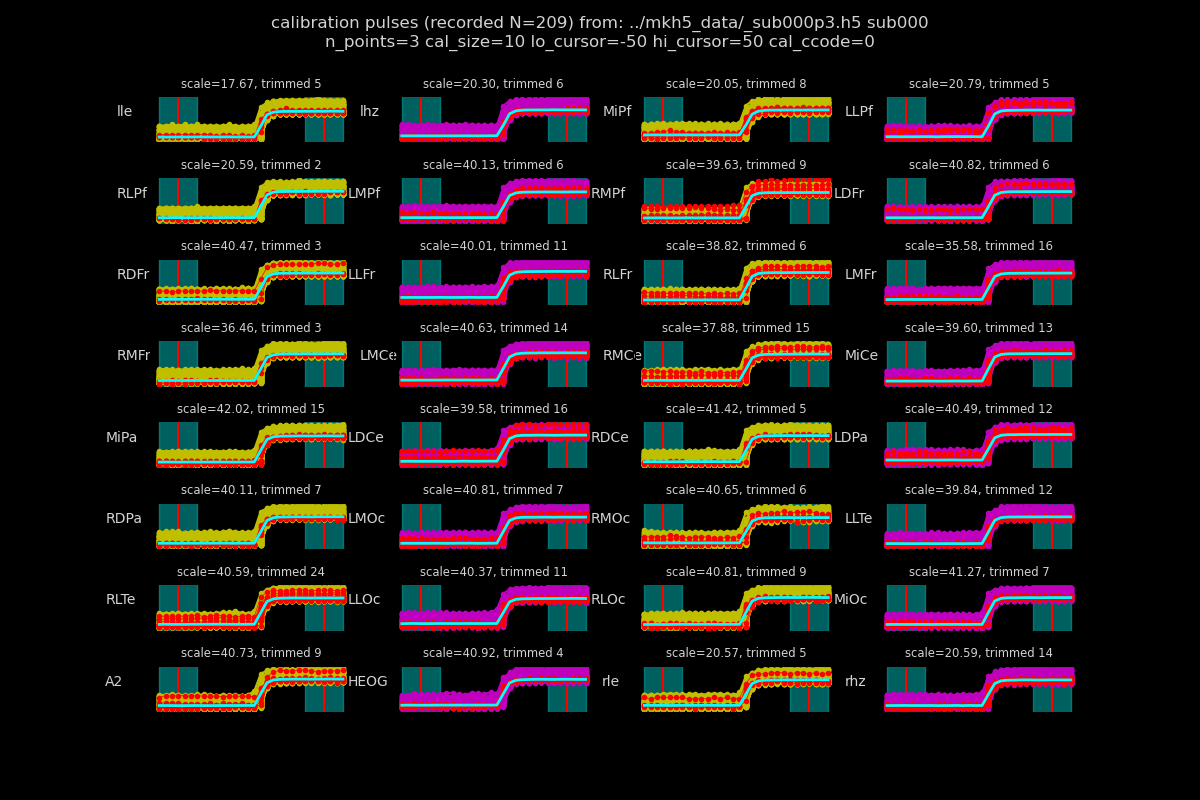
# Visually check the calibration parameters.
pts, pulse, lo, hi, ccode = 3, 10, -50, 50, 0
fig, ax = data_h5.plotcals(
DATA_H5_F,
"sub000",
n_points=pts, # pts to average, either side of cursor
cal_size=pulse, # uV
lo_cursor=lo, # lo_cursor ms
hi_cursor=hi, # hi_cursor ms
cal_ccode=ccode,
)
fig.show()
Out:
Plotting cals
/usr/share/miniconda/envs/env_3.8/lib/python3.8/site-packages/mkpy/mkh5.py:3666: UserWarning: negative event code(s) found for cal condition code 0 -16384
warnings.warn(msg)
Found cals in /sub000/dblock_4
Scale the EEG A/D to microvolts.
data_h5.calibrate_mkdata(
"sub000",
n_points=pts,
cal_size=pulse,
lo_cursor=lo,
hi_cursor=hi,
cal_ccode=ccode,
use_cals=None,
)
Out:
/usr/share/miniconda/envs/env_3.8/lib/python3.8/site-packages/mkpy/mkh5.py:3666: UserWarning: negative event code(s) found for cal condition code 0 -16384
warnings.warn(msg)
Found cals in /sub000/dblock_4
Calibrating block /sub000/dblock_0 of 5: (31232,)
Calibrating block /sub000/dblock_1 of 5: (32768,)
Calibrating block /sub000/dblock_2 of 5: (31744,)
Calibrating block /sub000/dblock_3 of 5: (32512,)
Calibrating block /sub000/dblock_4 of 5: (28416,)
Examine some header info with mkh5.headinfo()
data_h5.headinfo("dblock_0.*samplerate")
data_h5.headinfo("dblock_0.*MiPa")
Out:
sub000/dblock_0/samplerate: 250.0
sub000/dblock_0/apparatus/streams/MiPa/pos: MiPa
sub000/dblock_0/apparatus/streams/MiPa/neg: A1
sub000/dblock_0/apparatus/streams/MiPa/gain: 20000
sub000/dblock_0/apparatus/streams/MiPa/hphz: 0.01
sub000/dblock_0/apparatus/streams/MiPa/lphz: 100.0
sub000/dblock_0/apparatus/streams/MiPa/mne_type: eeg
sub000/dblock_0/apparatus/sensors/MiPa/x: 0.0
sub000/dblock_0/apparatus/sensors/MiPa/y: -2.7
sub000/dblock_0/apparatus/sensors/MiPa/z: 13.2
sub000/dblock_0/streams/MiPa/jdx: 23
sub000/dblock_0/streams/MiPa/source: dig_chan_0016
sub000/dblock_0/streams/MiPa/name: MiPa
sub000/dblock_0/streams/MiPa/dt: <f2
sub000/dblock_0/streams/MiPa/calibrated: True
sub000/dblock_0/streams/MiPa/cals/cal_srate: 250.0
sub000/dblock_0/streams/MiPa/cals/cal_dblock/0: ../mkh5_data/_sub000p3.h5/sub000/dblock_4
sub000/dblock_0/streams/MiPa/cals/cal_args/n_points: 3
sub000/dblock_0/streams/MiPa/cals/cal_args/cal_size: 10.0
sub000/dblock_0/streams/MiPa/cals/cal_args/lo_cursor: -50.0
sub000/dblock_0/streams/MiPa/cals/cal_args/hi_cursor: 50.0
sub000/dblock_0/streams/MiPa/cals/cal_args/cal_ccode: 0
sub000/dblock_0/streams/MiPa/cals/scale_by: 42.019882179675996
sub000/dblock_0/streams/MiPa/cals/var: 0.24502864170431732
sub000/dblock_0/streams/MiPa/cals/median: 42.0
sub000/dblock_0/streams/MiPa/cals/iqr: 0.7142857142857153
sub000/dblock_0/streams/MiPa/cals/n_cals: 194
sub000/dblock_0/streams/MiPa/cals/n_trimmed: 15
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/0: 27
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/1: 30
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/2: 35
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/3: 58
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/4: 68
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/5: 73
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/6: 76
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/7: 94
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/8: 101
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/9: 121
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/10: 159
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/11: 178
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/12: 181
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/13: 189
sub000/dblock_0/streams/MiPa/cals/trimmed_idxs/14: 206
Total running time of the script: ( 0 minutes 6.642 seconds)