pygarv: EEG viewer
pygarv is a command line utility that integrates EEG data
visualization with an EEG artifact screening test editor. It works
somewhat OK for visualizing mkh5 files, the artifact screening is work
in progress.
Panel dividers slide to hide or show the different views:
EEG artifact screening test editor
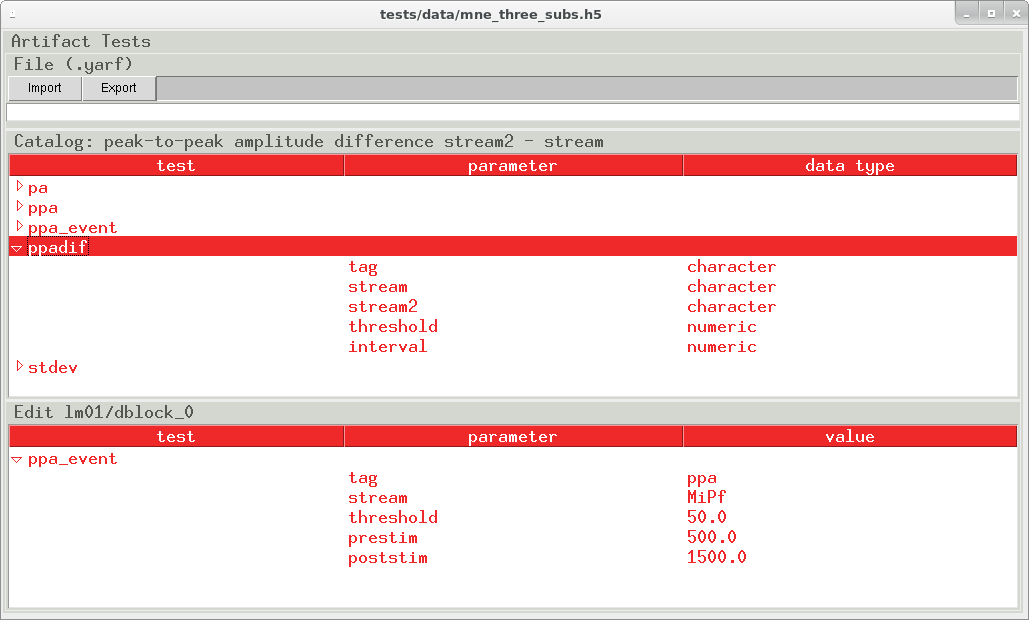
Artifact screening tests
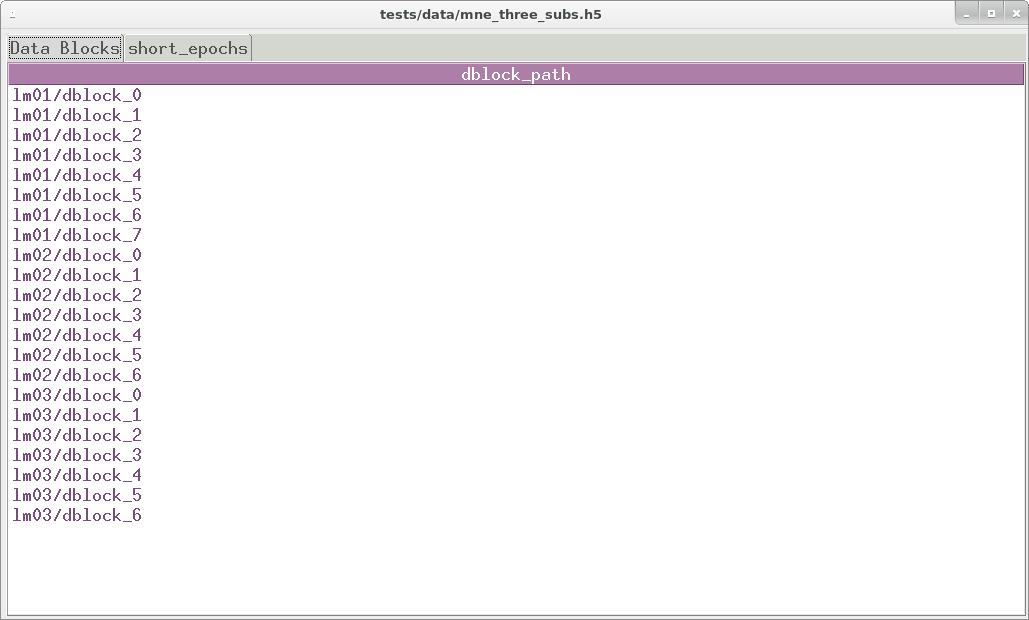
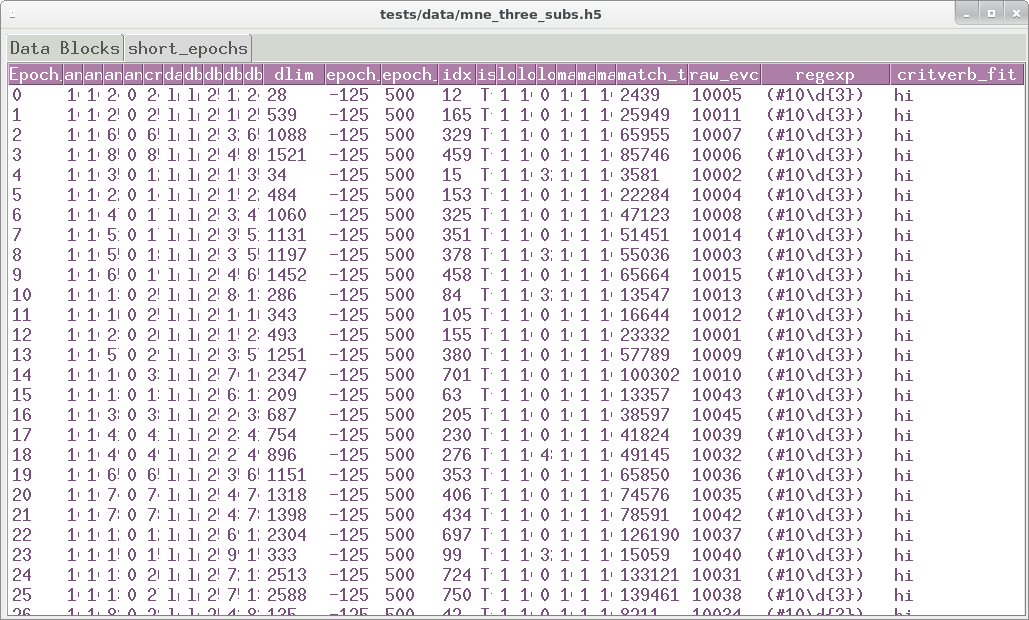
EEG artifacts in mkh5 files are tagged and tracked not deleted. A
dedicated pygarv 64-bit integer column alongside the EEG and event
codes is default 0 (== good) at every data block sample. A sequence of
user-specified Go-No-Go artifact tests is read from a YAML format text
file, stored in the data block header, and the tests swept across the
data. At samples where the n -th test fails, bit n of the
pygarv integer is set high. Any non-zero value in the pygarv
column indcates an artifact and the integer value can be decoded into
the indexes of the failed tests. In combination with the test
specifications stored in the header this can be use to reconstruct
exactly which tests failed at each sample. Since the pygarv column
travels with the EEG data, the artifact status of every data point is
available during subsequent analysis.
To streamline the EEG screening process, the pygarv dashboard integrates a test editor that can import, edit, visualize, and export the YAML files. This is a convenience, the YAML files can be generated programmatically or typed and edited by hand.