Note
Click here to download the full example code
append separate cals
Set up
from pathlib import Path
from matplotlib import pyplot as plt # for inspecting cal pulses
from mkpy import mkh5
MKDIG_PATH = Path("../mkdig_data")
print("mkpy version", mkh5.__version__)
Out:
mkpy version 0.2.7
Create subject, append cals
Use mkh5.append_mkdata() to
combine EEG recordings and separately recorded calibration data in
a single .h5 file prior to converting A/D samples to \(\mu\) V.
Set EEG and calibration data file names
crw = MKDIG_PATH / "sub000p3.crw" # EEG recording
log = MKDIG_PATH / "sub000p3.x.log" # events
yhdr = MKDIG_PATH / "sub000p3.yhdr" # extra header info
cals_crw = MKDIG_PATH / "sub000c.crw"
cals_log = MKDIG_PATH / "sub000c.log"
cals_yhdr = MKDIG_PATH / "sub000c.yhdr"
Convert .crw/.log to mkh5 format .h5.
p3_h5_f = "example_sub000p3.h5"
p3_h5 = mkh5.mkh5(p3_h5_f)
p3_h5.reset_all()
p3_h5.create_mkdata("sub000", crw, log, yhdr)
Append calibration data to the existing sub000.
p3_h5.append_mkdata("sub000", cals_crw, cals_log, cals_yhdr)
Convert EEG and calibration data A/D to microvolts. set re-usable calibration parameters
cal_kws = dict(
n_points=5, # samples to average
cal_size=10, # uV
lo_cursor=-36, # lo_cursor ms
hi_cursor=48, # hi_cursor ms
cal_ccode=0, # condition code
)
inspect the calibration pulses
f, ax = p3_h5.plotcals(p3_h5_f, "sub000", **cal_kws)
plt.show()
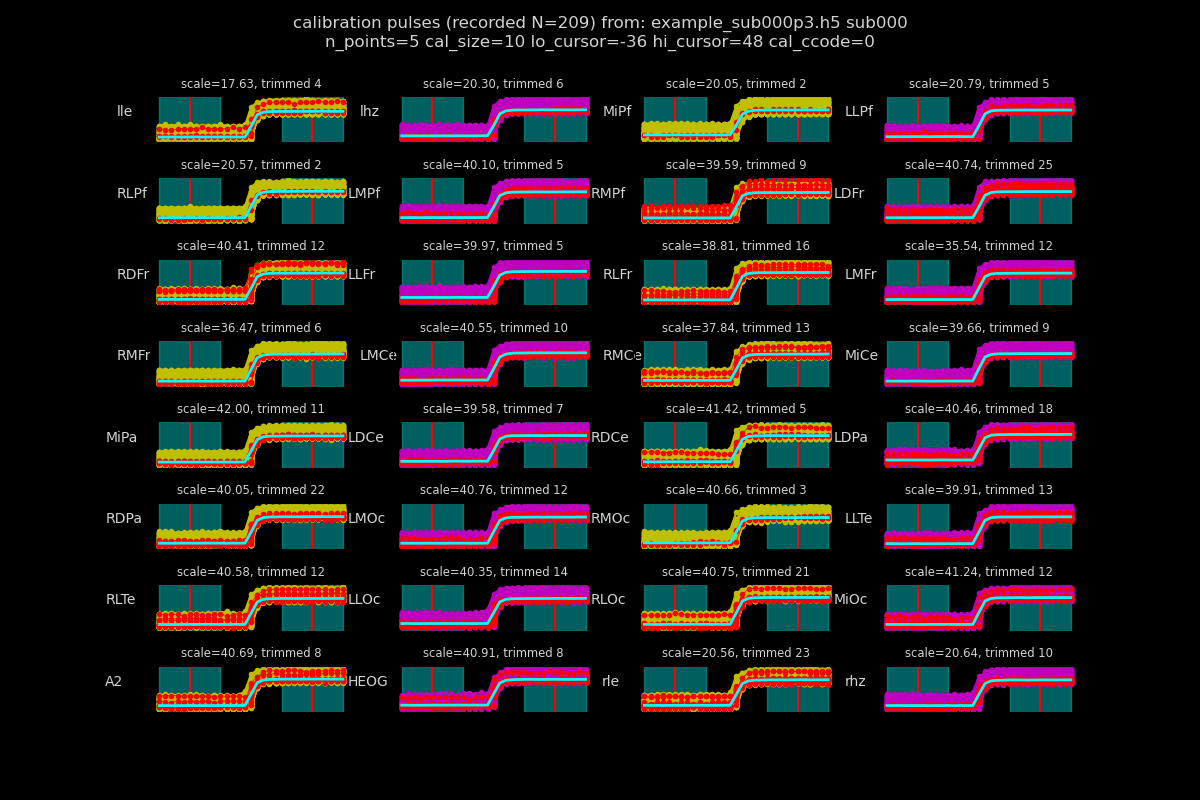
Out:
Plotting cals
/usr/share/miniconda/envs/env_3.8/lib/python3.8/site-packages/mkpy/mkh5.py:3666: UserWarning: negative event code(s) found for cal condition code 0 -16384
warnings.warn(msg)
Found cals in /sub000/dblock_4
Convert EEG and calibration data A/D to microvolts.
p3_h5.calibrate_mkdata("sub000", **cal_kws)
Out:
/usr/share/miniconda/envs/env_3.8/lib/python3.8/site-packages/mkpy/mkh5.py:3666: UserWarning: negative event code(s) found for cal condition code 0 -16384
warnings.warn(msg)
Found cals in /sub000/dblock_4
Calibrating block /sub000/dblock_0 of 5: (31232,)
Calibrating block /sub000/dblock_1 of 5: (32768,)
Calibrating block /sub000/dblock_2 of 5: (31744,)
Calibrating block /sub000/dblock_3 of 5: (32512,)
Calibrating block /sub000/dblock_4 of 5: (28416,)
Total running time of the script: ( 0 minutes 6.914 seconds)