Read single-trial EEG epochs
[1]:
import pandas as pd
from matplotlib import pyplot as plt
from spudtr import get_demo_df, P3_1500_FEATHER
from spudtr import epf
[2]:
epochs_df = get_demo_df(P3_1500_FEATHER)
# example: four midline EEG channels
eeg_channels = ['MiPf', 'MiCe', 'MiPa', 'MiOc']
# check the epochs format
epf.check_epochs(epochs_df, eeg_channels, epoch_id="epoch_id", time="time_ms")
# preview the entire dataframe
epochs_df
downloading ./spudtr/data/sub000p3.ms1500.epochs.feather from https://zenodo.org/record/3968485/files/ ... please wait
[2]:
| epoch_id | time_ms | sub_id | eeg_artifact | dblock_path | log_evcodes | log_ccodes | dblock_srate | ccode | instrument | ... | RMOc | LLTe | RLTe | LLOc | RLOc | MiOc | A2 | HEOG | rle | rhz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | -748 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -25.093750 | -0.753906 | 1.480469 | -13.414062 | -18.937500 | -17.734375 | 5.660156 | 98.875000 | -39.500000 | 38.375000 |
| 1 | 0 | -744 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -24.593750 | 0.502441 | -2.466797 | -17.640625 | -17.468750 | -15.304688 | 1.968750 | 104.750000 | -38.031250 | 41.281250 |
| 2 | 0 | -740 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -16.484375 | -1.507812 | 3.947266 | -15.648438 | -10.085938 | -11.171875 | 8.367188 | 102.062500 | -33.656250 | 43.718750 |
| 3 | 0 | -736 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -11.804688 | -15.070312 | 9.867188 | -14.906250 | -7.378906 | -8.742188 | 9.351562 | 100.562500 | -42.906250 | 37.406250 |
| 4 | 0 | -732 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -6.394531 | -4.019531 | 9.125000 | -10.679688 | -6.886719 | -8.015625 | 8.125000 | 98.375000 | -43.875000 | 37.906250 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 224995 | 600 | 732 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.671875 | -3.517578 | -4.441406 | -4.718750 | -4.671875 | -3.400391 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224996 | 600 | 736 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -4.195312 | -4.222656 | -4.425781 | -3.644531 | -4.429688 | -4.160156 | -3.412109 | -4.371094 |
| 224997 | 600 | 740 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -3.767578 | -4.441406 | -3.974609 | -4.425781 | -3.400391 | -4.429688 | -4.160156 | -3.900391 | -4.859375 |
| 224998 | 600 | 744 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -4.269531 | -4.195312 | -4.222656 | -4.425781 | -3.886719 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224999 | 600 | 748 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -3.947266 | -4.222656 | -4.179688 | -3.400391 | -4.183594 | -4.406250 | -3.412109 | -4.371094 |
225000 rows × 47 columns
Common A1 to average mastoid reference
a.k.a. “bimastoid”, “linked mastoid”
Warning, only valid for EEG recorded with a common A1 reference
[3]:
epf.re_reference(
epochs_df,
eeg_channels,
'A2',
"linked_pair",
epoch_id="epoch_id", time="time_ms",
)
[3]:
| epoch_id | time_ms | sub_id | eeg_artifact | dblock_path | log_evcodes | log_ccodes | dblock_srate | ccode | instrument | ... | RMOc | LLTe | RLTe | LLOc | RLOc | MiOc | A2 | HEOG | rle | rhz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | -748 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -25.093750 | -0.753906 | 1.480469 | -13.414062 | -18.937500 | -20.564453 | 5.660156 | 98.875000 | -39.500000 | 38.375000 |
| 1 | 0 | -744 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -24.593750 | 0.502441 | -2.466797 | -17.640625 | -17.468750 | -16.289062 | 1.968750 | 104.750000 | -38.031250 | 41.281250 |
| 2 | 0 | -740 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -16.484375 | -1.507812 | 3.947266 | -15.648438 | -10.085938 | -15.355469 | 8.367188 | 102.062500 | -33.656250 | 43.718750 |
| 3 | 0 | -736 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -11.804688 | -15.070312 | 9.867188 | -14.906250 | -7.378906 | -13.417969 | 9.351562 | 100.562500 | -42.906250 | 37.406250 |
| 4 | 0 | -732 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -6.394531 | -4.019531 | 9.125000 | -10.679688 | -6.886719 | -12.078125 | 8.125000 | 98.375000 | -43.875000 | 37.906250 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 224995 | 600 | 732 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.671875 | -3.517578 | -4.441406 | -4.718750 | -4.671875 | -1.185547 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224996 | 600 | 736 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -4.195312 | -4.222656 | -4.425781 | -1.429688 | -4.429688 | -4.160156 | -3.412109 | -4.371094 |
| 224997 | 600 | 740 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -3.767578 | -4.441406 | -3.974609 | -4.425781 | -1.185547 | -4.429688 | -4.160156 | -3.900391 | -4.859375 |
| 224998 | 600 | 744 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -4.269531 | -4.195312 | -4.222656 | -4.425781 | -1.671875 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224999 | 600 | 748 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -3.947266 | -4.222656 | -4.179688 | -1.308594 | -4.183594 | -4.406250 | -3.412109 | -4.371094 |
225000 rows × 47 columns
New common reference
Note: Only valid for common reference EEG data.
For example change from common A1 reference to a vertex or nose tip common reference.
Note: new the new reference = 0 as expected.
[4]:
# vertex location is MiCe
epf.re_reference(
epochs_df,
eeg_channels,
'MiCe',
"new_common",
epoch_id='epoch_id',
time='time_ms'
)
[4]:
| epoch_id | time_ms | sub_id | eeg_artifact | dblock_path | log_evcodes | log_ccodes | dblock_srate | ccode | instrument | ... | RMOc | LLTe | RLTe | LLOc | RLOc | MiOc | A2 | HEOG | rle | rhz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | -748 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -25.093750 | -0.753906 | 1.480469 | -13.414062 | -18.937500 | -20.515625 | 5.660156 | 98.875000 | -39.500000 | 38.375000 |
| 1 | 0 | -744 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -24.593750 | 0.502441 | -2.466797 | -17.640625 | -17.468750 | -11.257812 | 1.968750 | 104.750000 | -38.031250 | 41.281250 |
| 2 | 0 | -740 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -16.484375 | -1.507812 | 3.947266 | -15.648438 | -10.085938 | -7.882812 | 8.367188 | 102.062500 | -33.656250 | 43.718750 |
| 3 | 0 | -736 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -11.804688 | -15.070312 | 9.867188 | -14.906250 | -7.378906 | -6.212891 | 9.351562 | 100.562500 | -42.906250 | 37.406250 |
| 4 | 0 | -732 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -6.394531 | -4.019531 | 9.125000 | -10.679688 | -6.886719 | -12.062500 | 8.125000 | 98.375000 | -43.875000 | 37.906250 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 224995 | 600 | 732 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.671875 | -3.517578 | -4.441406 | -4.718750 | -4.671875 | 0.392578 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224996 | 600 | 736 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -4.195312 | -4.222656 | -4.425781 | 0.148438 | -4.429688 | -4.160156 | -3.412109 | -4.371094 |
| 224997 | 600 | 740 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -3.767578 | -4.441406 | -3.974609 | -4.425781 | 0.392578 | -4.429688 | -4.160156 | -3.900391 | -4.859375 |
| 224998 | 600 | 744 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -4.269531 | -4.195312 | -4.222656 | -4.425781 | -0.093750 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224999 | 600 | 748 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -3.947266 | -4.222656 | -4.179688 | 0.646484 | -4.183594 | -4.406250 | -3.412109 | -4.371094 |
225000 rows × 47 columns
Common average reference
Note: for demonstration only, a real application would use all scalp locations
[5]:
reference_channels = ["MiPf", "MiCe", "MiPa", "MiOc", "A2"]
epf.re_reference(
epochs_df,
eeg_channels,
reference_channels ,
"common_average",
epoch_id='epoch_id',
time='time_ms'
)
[5]:
| epoch_id | time_ms | sub_id | eeg_artifact | dblock_path | log_evcodes | log_ccodes | dblock_srate | ccode | instrument | ... | RMOc | LLTe | RLTe | LLOc | RLOc | MiOc | A2 | HEOG | rle | rhz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | -748 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -25.093750 | -0.753906 | 1.480469 | -13.414062 | -18.937500 | -3.210156 | 5.660156 | 98.875000 | -39.500000 | 38.375000 |
| 1 | 0 | -744 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -24.593750 | 0.502441 | -2.466797 | -17.640625 | -17.468750 | 1.857813 | 1.968750 | 104.750000 | -38.031250 | 41.281250 |
| 2 | 0 | -740 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -16.484375 | -1.507812 | 3.947266 | -15.648438 | -10.085938 | 2.100781 | 8.367188 | 102.062500 | -33.656250 | 43.718750 |
| 3 | 0 | -736 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -11.804688 | -15.070312 | 9.867188 | -14.906250 | -7.378906 | 3.550977 | 9.351562 | 100.562500 | -42.906250 | 37.406250 |
| 4 | 0 | -732 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -6.394531 | -4.019531 | 9.125000 | -10.679688 | -6.886719 | 0.596875 | 8.125000 | 98.375000 | -43.875000 | 37.906250 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 224995 | 600 | 732 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.671875 | -3.517578 | -4.441406 | -4.718750 | -4.671875 | 0.482812 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224996 | 600 | 736 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -4.195312 | -4.222656 | -4.425781 | 0.287500 | -4.429688 | -4.160156 | -3.412109 | -4.371094 |
| 224997 | 600 | 740 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -3.767578 | -4.441406 | -3.974609 | -4.425781 | 0.482812 | -4.429688 | -4.160156 | -3.900391 | -4.859375 |
| 224998 | 600 | 744 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -4.269531 | -4.195312 | -4.222656 | -4.425781 | 0.093750 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224999 | 600 | 748 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -3.947266 | -4.222656 | -4.179688 | 0.436719 | -4.183594 | -4.406250 | -3.412109 | -4.371094 |
225000 rows × 47 columns
Center EEG data in an interval (baseline)
The
startandstopinterval units are the same as the time channel
[6]:
start = -500
stop = -4
centered_eeg_df = epf.center_eeg(
epochs_df,
eeg_channels,
start,
stop,
epoch_id='epoch_id',
time='time_ms'
)
centered_eeg_df
[6]:
| epoch_id | time_ms | sub_id | eeg_artifact | dblock_path | log_evcodes | log_ccodes | dblock_srate | ccode | instrument | ... | RMOc | LLTe | RLTe | LLOc | RLOc | MiOc | A2 | HEOG | rle | rhz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | -748 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -25.093750 | -0.753906 | 1.480469 | -13.414062 | -18.937500 | -23.419617 | 5.660156 | 98.875000 | -39.500000 | 38.375000 |
| 1 | 0 | -744 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -24.593750 | 0.502441 | -2.466797 | -17.640625 | -17.468750 | -20.989929 | 1.968750 | 104.750000 | -38.031250 | 41.281250 |
| 2 | 0 | -740 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -16.484375 | -1.507812 | 3.947266 | -15.648438 | -10.085938 | -16.857117 | 8.367188 | 102.062500 | -33.656250 | 43.718750 |
| 3 | 0 | -736 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -11.804688 | -15.070312 | 9.867188 | -14.906250 | -7.378906 | -14.427429 | 9.351562 | 100.562500 | -42.906250 | 37.406250 |
| 4 | 0 | -732 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -6.394531 | -4.019531 | 9.125000 | -10.679688 | -6.886719 | -13.700867 | 8.125000 | 98.375000 | -43.875000 | 37.906250 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 224995 | 600 | 732 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.671875 | -3.517578 | -4.441406 | -4.718750 | -4.671875 | -3.422127 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224996 | 600 | 736 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -4.195312 | -4.222656 | -4.425781 | -3.666268 | -4.429688 | -4.160156 | -3.412109 | -4.371094 |
| 224997 | 600 | 740 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -3.767578 | -4.441406 | -3.974609 | -4.425781 | -3.422127 | -4.429688 | -4.160156 | -3.900391 | -4.859375 |
| 224998 | 600 | 744 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -4.269531 | -4.195312 | -4.222656 | -4.425781 | -3.908455 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224999 | 600 | 748 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -3.947266 | -4.222656 | -4.179688 | -3.422127 | -4.183594 | -4.406250 | -3.412109 | -4.371094 |
225000 rows × 47 columns
Exclude previously tagged artifacts
This special-purpose filter drops entire epochs where the time-locking event at time 0 is tagged as bad for some reason on the specified bads_column.
This implements a simple convention for pruning epochs based on tags generated by artifact screening functions.
Any column can be used or constructed for this purpose.
Example: drop all epochs where eeg_artifact is other than 0 at time_ms == 0
[7]:
good_epochs = epf.drop_bad_epochs(
epochs_df,
bads_column="eeg_artifact",
epoch_id='epoch_id',
time='time_ms',
)
print("Total number of epoch ids: ", len(epochs_df["epoch_id"].unique()))
print("Number of good epoch ids: ", len(good_epochs["epoch_id"].unique()))
good_epochs
Total number of epoch ids: 600
Number of good epoch ids: 542
[7]:
| epoch_id | time_ms | sub_id | eeg_artifact | dblock_path | log_evcodes | log_ccodes | dblock_srate | ccode | instrument | ... | RMOc | LLTe | RLTe | LLOc | RLOc | MiOc | A2 | HEOG | rle | rhz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | -748 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -25.093750 | -0.753906 | 1.480469 | -13.414062 | -18.937500 | -17.734375 | 5.660156 | 98.875000 | -39.500000 | 38.375000 |
| 1 | 0 | -744 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -24.593750 | 0.502441 | -2.466797 | -17.640625 | -17.468750 | -15.304688 | 1.968750 | 104.750000 | -38.031250 | 41.281250 |
| 2 | 0 | -740 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -16.484375 | -1.507812 | 3.947266 | -15.648438 | -10.085938 | -11.171875 | 8.367188 | 102.062500 | -33.656250 | 43.718750 |
| 3 | 0 | -736 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -11.804688 | -15.070312 | 9.867188 | -14.906250 | -7.378906 | -8.742188 | 9.351562 | 100.562500 | -42.906250 | 37.406250 |
| 4 | 0 | -732 | sub000 | 0 | sub000/dblock_0 | 0 | 0 | 250.0 | 1 | eeg | ... | -6.394531 | -4.019531 | 9.125000 | -10.679688 | -6.886719 | -8.015625 | 8.125000 | 98.375000 | -43.875000 | 37.906250 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 224995 | 600 | 732 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.671875 | -3.517578 | -4.441406 | -4.718750 | -4.671875 | -3.400391 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224996 | 600 | 736 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -4.195312 | -4.222656 | -4.425781 | -3.644531 | -4.429688 | -4.160156 | -3.412109 | -4.371094 |
| 224997 | 600 | 740 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -3.767578 | -4.441406 | -3.974609 | -4.425781 | -3.400391 | -4.429688 | -4.160156 | -3.900391 | -4.859375 |
| 224998 | 600 | 744 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.425781 | -4.269531 | -4.195312 | -4.222656 | -4.425781 | -3.886719 | -4.429688 | -4.406250 | -3.900391 | -4.371094 |
| 224999 | 600 | 748 | sub000 | 0 | sub000/dblock_4 | 0 | 0 | 250.0 | 0 | cal | ... | -4.179688 | -4.019531 | -3.947266 | -4.222656 | -4.179688 | -3.400391 | -4.183594 | -4.406250 | -3.412109 | -4.371094 |
203250 rows × 47 columns
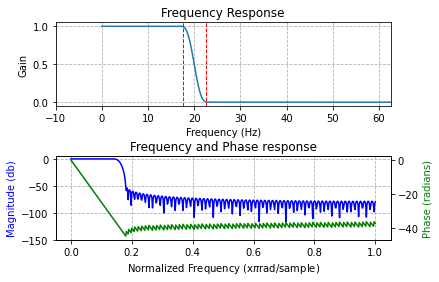
Filter EEG epochs:
Configure the filter
If left unspecified in show_filter() the optional parameters width_hz, ripple_db and window default to reasonable values.
However, to apply the filter all parameters must be explicitly specified. You can use the defaults or something else.
[8]:
from spudtr import filters
# note this fills in default transition width, ripple, and window values
lp_params = {
"ftype": "lowpass", # lowpass, bandpass, highpass
"cutoff_hz": 20, # 1/2 amplitude frequency Hz
"sfreq": 250.0, # sampling rate in samples/second
}
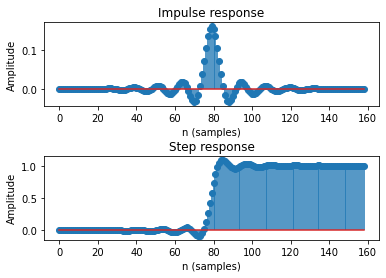
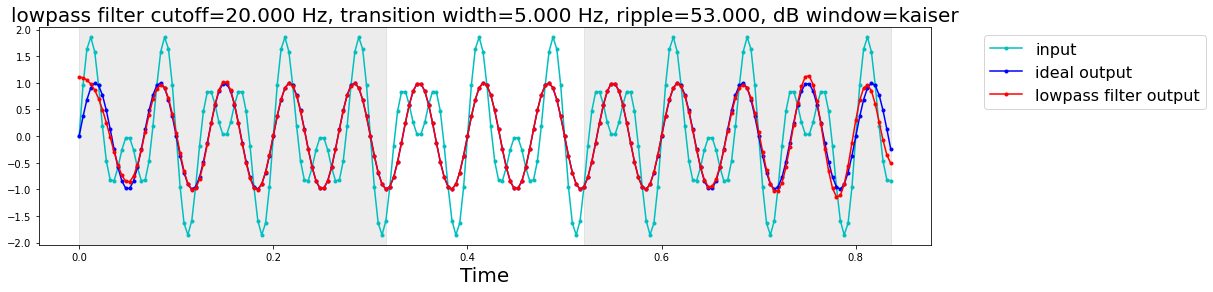
bode, imp, s_edge, n_edge = filters.show_filter(**lp_params); # expand the dictionary with python ** trick
lowpass filter
sampling rate (samples / s): 250.000
1/2 amplitude cutoff (Hz): 20.000
transition width (Hz): 5.000
ripple (dB): 53.000
window: kaiser
length (coefficients): 159
delay (samples): 79
edge distortion: first and last 0.3160 seconds of the data(= 79 samples at 250.0 samples / s)
/home/runner/miniconda/envs/env_3.9/lib/python3.9/site-packages/spudtr/filters.py:492: UserWarning: using default window='kaiser'
warnings.warn(f"using default window='{window}'")
/home/runner/miniconda/envs/env_3.9/lib/python3.9/site-packages/spudtr/filters.py:504: UserWarning: using default width_hz=5.000
warnings.warn(f"using default width_hz={width_hz:0.3f}")
/home/runner/miniconda/envs/env_3.9/lib/python3.9/site-packages/spudtr/filters.py:508: UserWarning: using default ripple_db=53.000
warnings.warn(f"using default ripple_db={ripple_db:0.3f}")
/home/runner/miniconda/envs/env_3.9/lib/python3.9/site-packages/spudtr/filters.py:885: UserWarning: linestyle is redundantly defined by the 'linestyle' keyword argument and the fmt string ".-" (-> linestyle='-'). The keyword argument will take precedence.
ax.plot(t, y, ".-", color="c", linestyle="-", label="input")
/home/runner/miniconda/envs/env_3.9/lib/python3.9/site-packages/spudtr/filters.py:886: UserWarning: linestyle is redundantly defined by the 'linestyle' keyword argument and the fmt string ".-" (-> linestyle='-'). The keyword argument will take precedence.
ax.plot(t, y1, ".-", color="b", linestyle="-", label="ideal output")
/home/runner/miniconda/envs/env_3.9/lib/python3.9/site-packages/spudtr/filters.py:887: UserWarning: linestyle is redundantly defined by the 'linestyle' keyword argument and the fmt string ".-" (-> linestyle='-'). The keyword argument will take precedence.
ax.plot(
Apply the filter
With all the parameters specified.
Note the **lp_params python trick to expand the filter parameter dictionary for the keyword arguments.
[9]:
# update the filter specs with default values
lp_params["window"] = "kaiser"
lp_params["width_hz"] = 5.0
lp_params["ripple_db"] = 53.0
display(lp_params)
epochs_df_lp = epf.fir_filter_epochs(
epochs_df,
data_columns=eeg_channels,
epoch_id="epoch_id",
time="time_ms",
trim_edges=False,
**lp_params,
)
times = epochs_df["time_ms"].unique()
epoch_ids = epochs_df["epoch_id"].unique()
{'ftype': 'lowpass',
'cutoff_hz': 20,
'sfreq': 250.0,
'window': 'kaiser',
'width_hz': 5.0,
'ripple_db': 53.0}
Compare the output
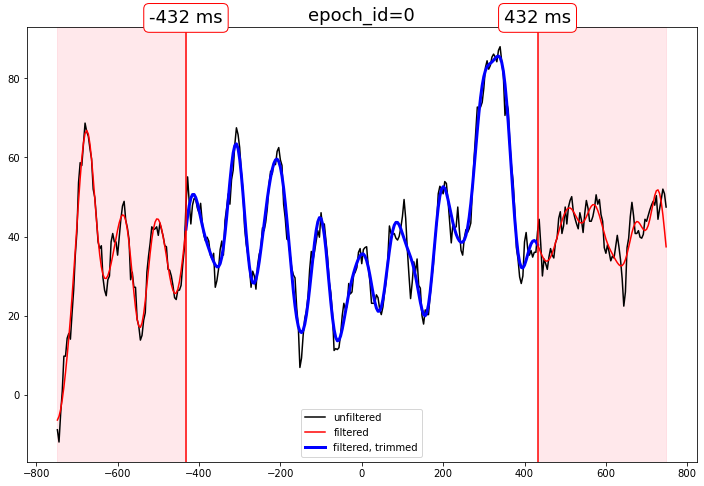
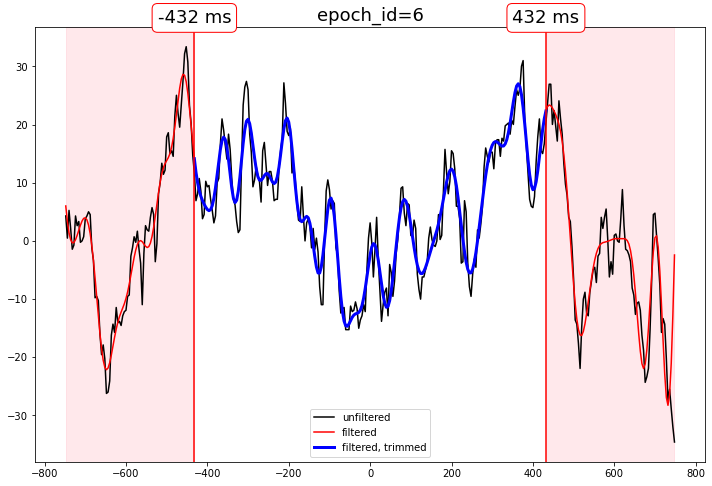
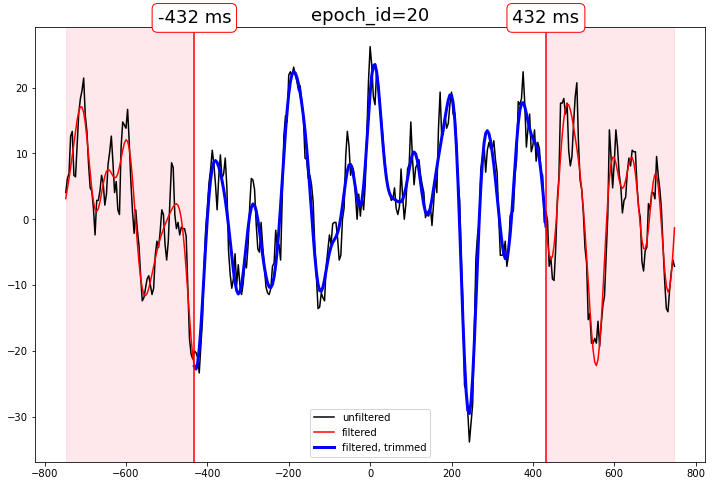
Filter EEG epochs, trim distorted edges
Edge distortion is reduced by zero-padding, but not eliminated.
How much to trim the epochs is your business.
[10]:
epochs_df_lp_trimmed = epf.fir_filter_epochs(
epochs_df,
data_columns=eeg_channels,
epoch_id="epoch_id",
time="time_ms",
trim_edges=True,
**lp_params,
)
trimmed_times = epochs_df_lp_trimmed["time_ms"].unique()
Compare the output
[11]:
# select some epochs to show
epidxs = [0, 6, 20]
channel = "MiPa"
for epidx in epidxs:
qstr = f"epoch_id == @epoch_ids[{epidx}]"
f, ax = plt.subplots(figsize=(12,8))
ax.set_title(f"epoch_id={epidx}", fontsize=18)
# unfiltered
ax.plot(
times,
epochs_df.query(qstr)[channel],
color="black",
label="unfiltered"
)
# filtered, phase compensated with distortion
ax.plot(
times,
epochs_df_lp.query(qstr)[channel],
color="red",
label="filtered"
)
# filtered, phase compensated, distortion trimmed
ax.plot(
trimmed_times,
epochs_df_lp_trimmed.query(qstr)[channel],
color="blue",
lw=3,
label="filtered, trimmed"
)
# decorate the beginning and end of the delay shift distortion regions
for xtime in [trimmed_times[0], trimmed_times[-1]]:
# beginning and end of the trimmed data
ax.axvline(xtime, color="red")
ax.annotate(
text=f"{str(xtime)} ms",
xy=(xtime, ax.get_ylim()[1]),
fontsize=18,
ha="center",
va="bottom",
bbox=dict(boxstyle="round", ec="red",fc="white")
)
# highlight the trimmed region
for bound in [0, -1]:
ax.axvspan(
times[bound],
trimmed_times[bound],
color="pink",
alpha=.2
)
ax.legend()